Biomed. adv. 1(1):12-19.
doi: 10.34172/bma.03
Review Article
Transcriptomic studies of cardiovascular biology and pathology in the light of human pluripotent stem cell-derived cardiomyocytes
Mojtaba Shafaghi Writing – original draft, 
Malihe Rezaee Writing – original draft,
Alireza Yaghoobi Writing – original draft,
Fatemeh Etezadi Writing – review & editing,
Sara Pahlavan Conceptualization, Supervision, Writing – review & editing, * 
Author information:
Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
Abstract
Summary
The biological science owes its recent advancement to the generation of in vitro human cellsthrough differentiation of pluripotent stem cells. This breakthrough not only helped to revisit many known cellular processes, but also founded the basis for discovery of many unknown and unanswered questions. In this regard, human induced pluripotent stem cells-derived cardiomyocyte (hiPSC-CM) played a pivotal role in high throughput transcriptomic studies of cardiac physiology and pathology. From the molecular pathways governing the development of cardiovascular system to the very far mechanistic studies of heart-related diseases, were identified using transcriptomes of in vitro generated human cardiomyocytes. This review summarizes the important steps toward turning hiPSC-CM to a valuable platform for high throughput transcriptomic studies and the major findings obtained in the light of using this tool.
Keywords: Human embryonic stem cell, Cardiomyocyte, Transcriptomics
Copyright and License Information
© 2024 The Author(s).
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Funding Statement
This work was supported by a grant from Royan Institute (Grant number: 400000195).
Introduction
Cell biology has long been the foundation of our understanding on the physiology and pathology of human body. It experienced some breakthroughs among which in vitro culture system might be considered the most influential one. Two-dimensional (2D) culture of primary cells and cell lines helped us to uncover various biological processes ranging from cell division to cell metabolism, apoptosis and much more in a controlled manner. However, primary cells experience senescence and cannot be sub-cultured infinitely. In contrast, cell lines are genetically engineered for escaping senescence and allow for infinite sub-culturing. While being very helpful, cell lines cannot represent the normal cellular states, as primary cells do. Furthermore, some primary cells are not feasible to access including cardiac or neural cells. Thus, there has been always a pressing need for unlimited access to these cells.
The isolation and in vitro culture of human embryonic stem cells (hESCs) in 1998 was a true turning point in the path toward having all human cell types in the lab.1 hESCs have unlimited proliferation capacity as well as differentiation potential towards all cell types in the human body.2 In the last three decades, cell biology has been equipped with optimal and advanced hESC culture conditions. Furthermore, differentiation protocols have been established for derivatives of all three embryonic germ layers; ectoderm, mesoderm and endoderm.3 Consequently, having human cardiac and neural cells in the lab was no longer a challenge. Cardiovascular research has experienced a breakthrough by employing cardiomyocytes differentiated from hESCs, at multiple levels of developmental, physiological and pathological studies. Advancements in cardiomyocyte differentiation protocols, higher efficiency and better reproducibility,4 have further helped the worldwide integration of this platform for cardiovascular research. Today, we are equipped with differentiation protocols for the generation of various cardiac lineage cells including cardiomyocytes, endothelial and smooth muscle cells as well as cardiac fibroblasts.5 Moreover, specific differentiation protocols have been introduced for various cardiomyocytes subtypes as well.6
The transcriptional signature of a cell determines its identity and function. Not so long ago, the focus of most subcellular studies was on coding RNAs. However, non-coding RNAs constitute the largest proportion of cellular RNAs with key regulatory functions.7 Therefore, investigating the bulk cellular RNA will provide a more realistic image of a cell condition. Advances in RNA screening techniques such as RNA sequencing (RNAseq) and making them more precise and specific, such as single cell RNAseq and single nucleus RNAseq, have provided an unprecedented opportunity to access the cellular transcriptional landscape. Combining the above-mentioned tools in cell biology has opened new horizons for cardiovascular research (Figure 1). We are now lucky to have access to an hESC-derived cardiomyocyte platform which helps uncover cardiac developmental mysteries when combined with transcriptomics, because of the access to stage-specific cells which could not have been accessed in a developing embryo. Furthermore, human cardiovascular diseases may be modeled in vitro by employing hESC-derived cardiomyocytes either through chemical/pharmacological interventions, or using patient-specific human induced pluripotent stem cells (hiPSCs)/genetically engineered isogenic hiPSCs. Mechanistic studies of cardiovascular diseases might become possible by integrating transcriptomic studies with in vitro human cardiac disease models. In this review, we highlight some achievements in cardiovascular biology, development and pathology by employing the transcriptomic studies with hESC-derived cardiomyocyte platforms.
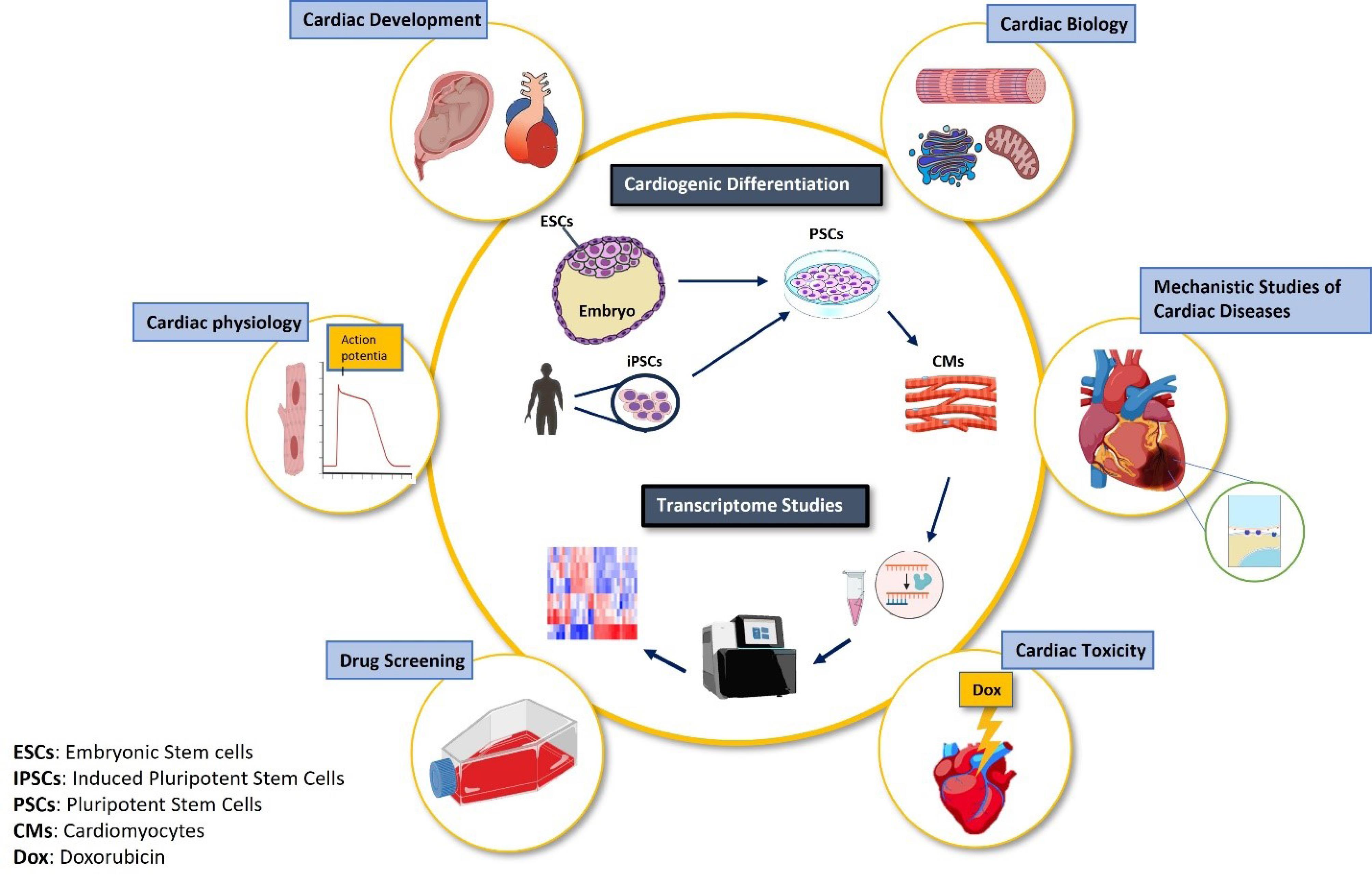
Figure 1.
A schematic illustration of advancement in cardiovascular research by employing combined in vitro cardiogenic differentiation platform with transcriptomic studies
.
A schematic illustration of advancement in cardiovascular research by employing combined in vitro cardiogenic differentiation platform with transcriptomic studies
Cardiac development
The heart is considered the first organ to be formed following embryonic gastrulation.8 The development of the heart starts from the anterior primitive streak,9 followed by cardiac mesodermal differentiation which occurs after the lateral movement of the anterior region into the splanchnic mesoderm.9 The cardiac mesoderm is specified through a sequence of events that involves the consecutive upregulation of specific transcription factors including Mesoderm Posterior BHLH Transcription Factor 1 (MESP-1), the surface markers kinase insert domain receptor (KDR) and platelet derived growth factor receptor (PDGFR).10 Then, the primary self-organized cardiac mesoderm divides into three regions including the first heart field (FHF), the second heart field (SHF) and the head fold.9 Cardiomyogenesis is then induced by the transcriptional activation of regulatory factors GATA4, TBX5 and SMARCD3 in the FHF.11 The adult heart consists of cardiomyocytes and a non-myocyte resident cell population including smooth muscle cells (SMCs), endothelial cells and cardiac fibroblasts. Cardiomyocytes are the most abundant cell population in the heart muscle, followed by cardiac fibroblasts and other cell types.12
In vitro differentiation of cardiomyocytes from human pluripotent stem cells
Human primary cells are an important source for biological and pathological studies; however, the isolation and subculture of certain primary cells remain an important challenge in cell biology to this day. human induced pluripotent stem cells (hiPSCs) have resolved a major part of these challenges due to their differentiation capacity into almost all cell types in human body, while retaining their unlimited proliferation capacity. Cardiovascular science has also benefited a lot from in vitro cardiogenesis using hiPSCs. Several protocols have been reported in an attempt to mimic the main signaling pathways that derive in vivo cardiogenesis.13 Thus, it is imperative to have a deep knowledge of cellular drivers of cardiac mesodermal, cardiac progenitor and cardiomyocyte specification in order to develop more efficient and specific protocols for in vitro differentiation. Furthermore, these principles can be applied to other cell types within cardiac tissue. Extensive research has been dedicated to unraveling the major cellular and molecular mechanisms of cardiogenesis using various cell and animal models over the last decades.14 As a result, cardiogenesis can be stimulated in vitro by following three steps: In the first step of hiPSC differentiation, it is necessary to modulate the three major TGFβ, BMP and canonical Wnt signaling pathways in order to promote cardiac mesoderm differentiation.15 The second step involves the specification of cardiac mesoderm cells towards cardiac progenitor cells (CPCs), in which inhibition of Wnt signaling pathways is required.15 Studies have shown that TGFβ and Wnt signaling have a biphasic effect on cardiomyogenesis. Downregulation of TGFβ promotes the differentiation of CPCs towards cardiomyocytes,15 while a sustained and continued TGFβ signaling pushes the cells toward the vascular smooth muscle and endothelial lineages and distracting them from the cardiomyocyte (CM) fate.15 In the last step of differentiation, mature CMs are formed. Several cardiac transcription factors, including NKX2,5, TBX5 and GATA4, cooperate to induce transcription of cardiac structural and excitation-contraction coupling genes.15
The in vitro differentiation of hiPSCs into cardiac lineage cells can be promoted in traditional two-dimensional (2D) monolayers or by using three-dimensional (3D) embryoid bodies through temporal regulation of the Activin/Nodal, BMP and Wnt signaling pathways.15 One of the widely used protocols is the GiWi (GSK-inhibition/Wnt inhibition), which enables the generation of about 80% CMs in a highly reproducible manner. The major event is the generation of cardiac mesodermal precursors by inhibition of glycogen synthase kinase-3β (GSK-3β) and the canonical Wnt signaling.16 It should be mentioned that despite enormous progress, in vitro-generated cardiomyocytes have an immature phenotype and lack some important characteristics of an adult cardiomyocyte. hiPSC-derived cardiomyocytes (hiPSC-CM) have a round shape, lack t-tubules and a mature Ca2+ handling system, and have undeveloped excitability due to a lack of some K + channels, and contain less mitochondria, all of which make hiPSC-CM very similar to fetal CMs.17 Therefore, major attempts have been put into the development of CM maturation strategies using several approaches, individually or together, by specifically targeting electro-mechanics, metabolism and tissue construction.18
Enlightening the cardiovascular biology by transcriptomic studies
Our knowledge of cardiovascular biology has experienced enormous progress with the introduction of innovative omics technologies. Particularly, we have obtained comprehensive information on the molecular regulators of all cellular events at multiple levels of genomics, transcriptomics, and proteomics. While we previously had an overview of specific gene expression profiles during important biological processes such as the cell cycle, development and apoptosis, our understanding of total RNA dynamics has taught us new aspects of each cellular event and the related regulatory mechanisms. This has helped us to unravel disease mechanisms which were a black box throughout human lifetime. Especially, non-coding RNAs have opened a totally new horizon in the understanding of gene regulatory networks as the basis for normal and abnormal biological processes. With respect to cardiovascular development, cardiomyocyte physiology and cardiovascular disease mechanisms, transcriptomic studies have empowered the field to a large extent, so that we are now well equipped with the necessary information to develop novel therapeutic strategies.
Decoding the molecular mechanisms of cardiovascular development by total RNA screening
RNA molecules exist in various forms and functions in the cell, with the non-coding ones being the most frequent.19 RNA sequencing technology enabled us to study both coding and non-coding RNAs simultaneously and obtain valuable information regarding gene expression and regulation at the same time. Our knowledge of molecular mechanisms of cardiac development has been highly influenced by the screening of total cellular RNA. It should be noted that a platform including cardiomyocytes differentiated from hiPSCs, combined with RNA sequencing, offers unprecedented opportunities for developmental studies. This is because in vitro cardiogenesis enables a time series investigation of heart development without the challenges of sampling the embryonic heart and related ethical issues. Indeed, multiple studies have taken advantage of these combined technologies and obtained remarkable findings. The miRNA expression profile was investigated in cardiac progenitors and early-differentiated cardiomyocytes by applying an NKX2,5 (eGFP/w) hiPSC reporter line. Interestingly, the canonical myogenic miRNAs (MIR1-1, MIR133A1 and MIR208A) were enriched in cardiomyocytes, while the NKX2,5-dependent gene regulatory network was not involved, either in mesoderm commitment or in cardiomyocyte specification. Therefore, the cardiac myogenic miRNA program is highlighted in early human cardiomyocyte commitment and differentiation, independent of the highly conserved NKX2,5-dependent transcriptions.20 In addition to the myogenic miRNA program, the expression level of all MIR148A family members markedly increased during the differentiation of hESCs into cardiomyocytes.21 In the absence of the MIR148A family (MIR148A-TKO), the cardiomyocytes yield from in vitro hESC differentiation was substantially decreased, which was rescued by the ectopic expression of MIR148A family members. Further gene expression profiling showed a linear correlation between the MIR148A family members upregulation and lateral mesoderm differentiation at the expense of paraxial mesodermal fate from primitive streak cells. This function has been attributed to the NOTCH ligand Delta-like 1 (DLL1) as the target gene of the MIR148A family, where DLL1 knockdown could enhance the directed differentiation of MIR148A-TKO hESCs into cardiomyocytes.21 This study identified the MIR148A family members as prospective regulators of cardiogenesis by counterbalancing mesoderm lineage commitment from paraxial to lateral mesoderm commitment.
While the molecular mechanisms of heart development have become less mysterious by taking advantage of in vitro hESCs differentiation and gene expression profiling, single cell RNA sequencing has provided even more knowledge on the spatiotemporal resolution of cardiovascular lineage cells during cardiogenesis. In an attempt to unravel lineage specification of the FLK+ mesoderm and its underlying mechanism, it has been shown that the smooth muscle lineage maintains the transcriptional signature of the FLK+ mesoderm, suggesting this mesoderm as its default ancestor.22 Interestingly, transcriptome analysis enabled a precise correlation of in vitro differentiation stages with the human embryonic heart developmental window.23 hESC-derived CPCs showed the most overlap with the gene expression profile of human embryonic hearts during Carnegie stage (CS) 10. Concordantly, the gene expression profile of hESC-derived cardiomyocytes was highly correlated with embryonic hearts at CS14-CS16. This finding strengthened the possibility of using in vitro cardiomyocyte differentiation platform for cardiac developmental studies.
In addition to the myogenic lineage, hESC-derived CPCs have the potency to generate nonmyogenic lineages such as cardiac fibroblast-like cells and endothelial cells. When these progenitor cells were characterized at a single-cell resolution and subjected to trajectory mapping in order to identify the branching points of derivatives, the presence of nonmyogenic populations was validated along with their particular markers such as TGFBI for cardiac fibroblasts. ISL1 was shared between both myogenic and nonmyogenic derivatives, which classified it as an early-stage multipotent cardiac progenitor marker. Interestingly, the knockdown of ISL1 in myogenic progenitors redirected them towards neural-like populations. Stage-specific comparison of myogenic progenitors’ differentiation with the embryonic human heart transcriptome at 5-week, highlighted the larger distance between early progenitors and the 5-week embryonic heart, while being less distant from late-stage progenitor cells. Therefore, time-series developmental studies are possible with this in vitro platform, while in vivo samples in some early stages are not accessible.24
In addition to a widely accepted platform for cardiac developmental studies, hESC-derived cardiomyocytes have been applied to investigate the conserved mechanisms of cardiac development between mice and humans. As the majority of our knowledge of heart development comes from in vivo studies using transgenic mouse models, the existing data should be examined in humans as well. In vitro cardiomyocytes differentiation has equipped us with a relevant model for such investigations. For instance, the determinative role of Asb2 (Cullin5 Ub ligase key subunit) in embryonic heart survival and complete looping was identified in a conditional AHF-Cre.Asb2 knockout mouse. CRISPR/Cas9 engineered hESCs validated the role of this ubiquitin ligase in in vitro cardiomyocyte differentiation, suggesting a conserved cardiac developmental mechanism.25 Furthermore, the developmental trajectory of the FHF and SHF was simulated in vitro by employing a double reporter hESC line (FHF: TBX5 ( + ) NKX2-5 ( + ) and SHF: TBX5 (-) NKX2-5 ( + )).26 Cardiomyocytes derived from engineered hESCs were analyzed on multiple levels of cellular, molecular and functional characteristics, which resulted in valuable findings regarding their ultimate cell population shares in the cardiac multicellular structure. When both bulk RNA and single cell RNA (scRNA) were employed, TBX5 expression was identified as a discriminative point for sarcomeric structure, oxidative phosphorylation, and calcium ion handling instruction. FHF-derived cardiomyocytes showed higher degree of maturity with respect to sarcomeric organization, larger ATP generation-linked oxygen consumption rate and excitation-contraction coupling components compared to SHF-derived cardiomyocytes. These characteristics placed FHF-derived cardiomyocytes in closer association with human fetal cardiomyocytes in the differentiation trajectory, which was drawn by pseudotime analyses. Another attempt to categorize cardiac progenitors, focused on the comparison of the hiPSC-derived and mouse mesoderm transcriptomes, which succeeded in the identification of three distinct human FHF, anterior SHF (aSHF), and posterior SHF (pSHF) mesoderm subpopulations.27
Inspired by signaling pathways identified from transcriptomics, in vitro cardiomyocyte differentiation was instructed to generate cardiomyocyte subtypes. These findings further unveiled the molecular mechanisms underlying cardiomyocyte subtypes specification and whole heart formation. Interestingly, combining single-cell transcriptomics with genetic labelling and ex vivo human-mouse embryonic chimeras resulted in the identification of retinoic acid signaling as a key regulator of heart field progenitors’ specification.28 This multi-angled analysis uncovered a new population of cardiac field progenitors that differentiated into both myocardial and epicardial cells. Applying these principles to in vitro mechanistic studies of congenital heart diseases, specific transcriptional dysregulation of first and SHF progenitors was observed in cardiogenic differentiation of patient-specific induced pluripotent stem cells (iPSCs) with hypoplastic left heart syndrome. These findings further validate the in vitro cardiomyocyte differentiation platform, not only for cardiac developmental studies, but also for human cardiac disease modeling. In continuation to uncovering early cardiac development, HAND1/2-/- NKX2.5(eGFP) H9 human embryonic stem cells were differentiated into cardiomyocytes and subjected to transcriptomic studies.29 Interestingly, single HAND1 or HAND2 knockout did not influence the differentiation kinetics, while a double knockout delayed the differentiation onset. HAND1 and HAND1/2 knock out changed the balance of progenitor population towards SHF and its derivatives, whereas HAND2 knockout was more associated with a higher incidence of FHF-derived lineage of ventricular cardiomyocytes. Further analysis identified TBX5 as one of the downstream targets of HAND1/2, which overexpression partially rescued the unbalanced cardiomyocyte differentiation in HAND1/2-knockout hESCs.
Cardiomyocytes’ myofibrillogenesis is a crucial step in heart development due to its fundamental role in contractile function. Various isoforms of sarcomeric proteins resulting from splicing variants, promote developmentally-distinct stages of myofibrillogenesis. This process was precisely investigated by using a CRISPR/Cas9 engineered RBM24(-/-) hESC.30 As RBM24 (RNA-binding motif protein 24) is a tissue-specific splicing regulator, its absence influenced sarcomeric organization and resulted in punctate Z-lines due to impaired myosin replacement during cardiomyocyte differentiation. Gene expression profiling of RBM24(-/-) hESC-derived cardiomyocytes identified over 4000 differentially expressed genes among which splice variants of core myofibrillogenesis proteins were abundant. For instance, ACTN2 [α-actinin 2], TTN [titin], and MYH10 [non-muscle myosin IIB]) were mis-regulated, as a consequence, muscle myosin isoforms such as MYH6 (muscle myosin II) could not replace MYH10 as a non-muscle myosin isoform and myofibrillogenesis was impaired. With respect to ACTN2, a splice variant lacking the ABD (actin-binding domain; encoded by exon 6) was upregulated in early cardiomyocytes derived from the differentiation of RBM24(-/-) hESCs, which further promoted sarcomere disassembly. Therefore, the in vitro cardiomyocyte differentiation platform validated the regulatory role of RBM24 in temporal dynamics of core myofibrillogenesis genes for sarcomere organization.
Empowering the cell and molecular biology of cardiomyocytes by transcriptomic studies of in vitro generated human cardiac cells
The excitation-contraction coupling of cardiomyocytes establishes the foundation of the heart pumping function and is crucial to normal human physiology. While cellular components of this process have been majorly identified, there are still unknown aspects that need to be elucidated. In vitro cardiogenesis provides an unprecedented opportunity to unravel the precise mechanisms of this key physiological process. Additionally, this platform facilitates the study of other cell biology attributes such as transcription, translation, cell cycle, apoptosis, autophagy and much more. In this regard, RYBP (epigenetic regulator RING1 and YY1 binding protein) has been suggested as an essential component of contractility in hESC-derived cardiomyocytes.31 To further identify the precise molecular mechanism, the dynamic transcriptome profile of the differentiating RYBP null mutant hESCs was compared to their wild-type counterparts. Interestingly, RYBP was broadly expressed in progenitor cells, while downregulating in the terminally differentiated wild-type hESC-derived cardiomyocytes. RYBP knockout resulted in the differential expression of genes related to ion homeostasis, cell adhesion, and sarcomeric organization. Of note, it seems that RYBP deletion results in These transcriptional dysregulations secondary to its role in the proper expression of key cardiac transcription factors including MESP1, SHH and MEF2C. Cardiac development involves a critical step of a metabolic shift from glycolysis to oxidative phosphorylation. This metabolic paradigm change requires mitochondrial development, functionalization and abundance in mature cardiomyocytes. Using transcriptome analysis of differentiating iPSCs, the developmental dynamism and subcellular mechanism of mitochondrial energy metabolism (MEM) were explored.32 A screening of differentially expressed genes was performed with a particular focus on the upregulated MEM genes during cardiac lineage commitment, which identified CCK and NOS3. Furthermore, some upregulated MEM genes and their anticipated protein products were related to cardiomyocyte maturation through their interaction with cardiac muscle contractile proteins. Thus, MEM was suggested as a key regulatory component of cardiomyocyte maturation. In addition to cellular processes, the in vitro cardiomyocyte differentiation platform facilitates subcellular studies such as post-transcriptional regulation and translational control.33 This was the objective of a study where polysome profiling followed by RNA sequencing was used to identify the transcripts showing dynamic 3’ UTR lengthening or shortening along with active recruitment to ribosome complexes, during cardiogenic differentiation of hiPSCs.33 The findings indicated that while the pluripotent stage had a preference for longer 3’ UTRs, cardiomyocyte commitment coincided with a preference for shorter 3’ UTRs. Mesoderm specification was marked as the stage with the most distinct regulatory changes.
Upgrading the mechanistic studies of cardiovascular diseases by transcriptomic analysis of in vitro cardiogenesis
Patient-specific iPSCs and in vitro cardiogenic differentiation, paved the way for uncovering the disease mechanisms in a controlled and time-dependent manner. Mitochondrial trifunctional protein deficiency which results in sudden infant death syndrome with no cure, was modeled using hydratase subunit A (HADHA)-deficient hiPSCs.34 These hiPSCs were subjected to cardiomyogenesis and accelerated maturation by an engineered microRNA maturation cocktail. Fatty acid treatment of matured HADHA-mutant cardiomyocytes recapitulated the disease phenotype including abnormal repolarization and calcium dynamics resulting in a pro-arrhythmic condition. Single cell RNA sequencing revealed a dysregulated profile of metabolic genes expression during cardiomyocyte differentiation of HADHA-deficient hiPSCs. This transcriptional change resulted in reduced fatty acid beta-oxidation, reduced mitochondrial proton gradient, disrupted cristae structure and defective cardiolipin remodeling, which all suggest a crucial role for HADHA in normal mitochondrial function.
QKI which is an RNA-binding protein and a well-known regulator of pre-mRNA alternative splicing, is highly expressed in developing and adult hearts. QKI-deficient hESCs (hESCs-QKI(del)) were differentiated into cardiomyocytes and were studied with respect to differentiation capacity and transcriptomic profile.35 While mesoderm induction and cardiac progenitor specification was normal in hESCs-QKI(del), functional cardiomyocytes were not obtained. Further transcriptional and functional analyses suggested an impaired sarcomerogenesis through dysregulation of alternative splicing in genes involved in Z-disc formation. Thus, QKI was proposed as a key molecule for certain types of cardiomyopathies.
While myomesin-1 (MYOM1)-related myopathies were lifelong uncharacterized disorders due to the lack of a knockout mouse model, MYOM1 knockout hESCs (MYOM1(-/-) hESC) enabled the study of the underlying mechanism. MYOM1(-/-) hESC-derived cardiomyocytes recapitulated the myocardial atrophy phenotype and uncovered the role of myomesin-1 in cardiomyocytes development, sarcomere assembly and contractility regulation.36 Therefore, this platform was validated as a disease model which can be used for studying the pathophysiology and treatment strategies.
The homeodomain transcription factor SHOX2 is a key molecule in cardiac conduction-related diseases such as atrial fibrillation and sinus node dysfunction. Its primary role in sinoatrial node (SAN) development explains the pathological association. The establishment of a Shox2(-/-) mouse ESCs facilitated the mechanistic studies of Shox involvement in SAN development and disease pathogenesis. The bulk RNAseq of Shox2(-/-) and Shox2( + / + ) mESC-derived SAN-like cells identified 94 differentially expressed genes, among which 5 novel identified shox2 target genes were assigned a role in specification of conduction traits. These target genes included Cav1, Fkbp10, Igfbp5, Mcf2l and Nr2f2.37
Another important mechanistic study employing an hESC-derived cardiomyocyte platform uncovered the molecular pathway of an inherited cardiomyopathy related to the human L39X phospholamban (PLN) mutant.38 While L39X was introduced as a null mutation, the resulting cardiomyopathy was associated with a complete lack of detectable protein. To resolve the underlying mechanism, a panel of mutant and wild-type PLN modified mRNA (modRNA) constructs was designed and transfected into hESC-derived cardiomyocytes. Chemical inhibition of the Proteasomal complex in cardiomyocytes transfected with L39X mutant PLN modRNA resulted in a marked increase in protein expression levels. Transcriptional profiling of the wildtype and mutant PLN transfected hESC-derived cardiomyocytes identified a distinct gene expression signature for protein degradation pathways. Therefore, the proteasomal pathways were suggested as key mechanism for the development of the cardiomyopathic PLN null mutant L39X phenotype.
Perspective and future directions
Despite the astonishing results, the 2D culture of a single cardiac cell type might not fully mimic the cardiac tissue condition for cardiovascular biology, development, disease modeling and drug screening studies. 3D cultures such as cardiac organoid, cardiac microtissue and engineered heart tissue may better serve the above-mentioned purposes due to having a complete spectrum of cardiac tissue cell types, required cell-cell and cell matrix interactions as well as appropriate maturity. Therefore, attempts should be directed towards establishing proper 3D cardiac tissue-like structures with the highest degree of similarity to native cardiac tissue.
Conclusion
High throughput omics studies of cardiac biology and pathology requires a platform with two distinct properties; (1) Human cardiac cells and (2) Large quantity of cells. Therefore, hiPSC-CM might be the best option for these studies due to providing both aforementioned properties. Advances in culture and differentiation methods would definitely help to turn this platform to a widely used tool for such studies in the near future.
Competing Interests
The authors declare no conflict of interest.
Ethical Approval
Not applicable.
Acknowledgements
The authors would like to thank Royan Cardiovascular Group for their help and support.
References
- Liu G, David BT, Trawczynski M, Fessler RG. Advances in pluripotent stem cells: history, mechanisms, technologies, and applications. Stem Cell Rev Rep 2020; 16(1):3-32. doi: 10.1007/s12015-019-09935-x [Crossref] [ Google Scholar]
- Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K. Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays. Biomaterials 2010; 31(15):4360-6. doi: 10.1016/j.biomaterials.2010.02.012 [Crossref] [ Google Scholar]
- Priester C, MacDonald A, Dhar M, Bow A. Examining the characteristics and applications of mesenchymal, induced pluripotent, and embryonic stem cells for tissue engineering approaches across the germ layers. Pharmaceuticals (Basel) 2020; 13(11):344. doi: 10.3390/ph13110344 [Crossref] [ Google Scholar]
- Lyra-Leite DM, Gutiérrez-Gutiérrez Ó, Wang M, Zhou Y, Cyganek L, Burridge PW. A review of protocols for human iPSC culture, cardiac differentiation, subtype-specification, maturation, and direct reprogramming. STAR Protoc 2022; 3(3):101560. doi: 10.1016/j.xpro.2022.101560 [Crossref] [ Google Scholar]
- Kahn-Krell A, Pretorius D, Guragain B, Lou X, Wei Y, Zhang J. A three-dimensional culture system for generating cardiac spheroids composed of cardiomyocytes, endothelial cells, smooth-muscle cells, and cardiac fibroblasts derived from human induced-pluripotent stem cells. Front Bioeng Biotechnol 2022; 10:908848. doi: 10.3389/fbioe.2022.908848 [Crossref] [ Google Scholar]
- Afjeh-Dana E, Naserzadeh P, Moradi E, Hosseini N, Seifalian AM, Ashtari B. Stem cell differentiation into cardiomyocytes: current methods and emerging approaches. Stem Cell Rev Rep 2022; 18(8):2566-92. doi: 10.1007/s12015-021-10280-1 [Crossref] [ Google Scholar]
- Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol 2023; 24(6):430-47. doi: 10.1038/s41580-022-00566-8 [Crossref] [ Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 2005; 6(11):826-35. doi: 10.1038/nrg1710 [Crossref] [ Google Scholar]
- Thomas D, Choi S, Alamana C, Parker KK, Wu JC. Cellular and engineered organoids for cardiovascular models. Circ Res 2022; 130(12):1780-802. doi: 10.1161/circresaha.122.320305 [Crossref] [ Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med 2000; 10(8):345-52. doi: 10.1016/s1050-1738(01)00069-x [Crossref] [ Google Scholar]
- Feulner L, van Vliet PP, Puceat M, Andelfinger G. Endocardial regulation of cardiac development. J Cardiovasc Dev Dis 2022; 9(5):122. doi: 10.3390/jcdd9050122 [Crossref] [ Google Scholar]
- Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 2014; 114(3):511-23. doi: 10.1161/circresaha.114.300558 [Crossref] [ Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 2011; 8(2):228-40. doi: 10.1016/j.stem.2010.12.008 [Crossref] [ Google Scholar]
- Lamberto F, Peral-Sanchez I, Muenthaisong S, Zana M, Willaime-Morawek S, Dinnyés A. Environmental alterations during embryonic development: studying the impact of stressors on pluripotent stem cell-derived cardiomyocytes. Genes (Basel) 2021; 12(10):1564. doi: 10.3390/genes12101564 [Crossref] [ Google Scholar]
- Doyle MJ, Lohr JL, Chapman CS, Koyano-Nakagawa N, Garry MG, Garry DJ. Human induced pluripotent stem cell-derived cardiomyocytes as a model for heart development and congenital heart disease. Stem Cell Rev Rep 2015; 11(5):710-27. doi: 10.1007/s12015-015-9596-6 [Crossref] [ Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD. Chemically defined generation of human cardiomyocytes. Nat Methods 2014; 11(8):855-60. doi: 10.1038/nmeth.2999 [Crossref] [ Google Scholar]
- Correia C, Christoffersson J, Tejedor S, El-Haou S, Matadamas-Guzman M, Nair S. Enhancing maturation and translatability of human pluripotent stem cell-derived cardiomyocytes through a novel medium containing acetyl-CoA carboxylase 2 inhibitor. Cells 2024; 13(16):1339. doi: 10.3390/cells13161339 [Crossref] [ Google Scholar]
- Hong Y, Zhao Y, Li H, Yang Y, Chen M, Wang X. Engineering the maturation of stem cell-derived cardiomyocytes. Front Bioeng Biotechnol 2023; 11:1155052. doi: 10.3389/fbioe.2023.1155052 [Crossref] [ Google Scholar]
- Bhatti GK, Khullar N, Sidhu IS, Navik US, Reddy AP, Reddy PH. Emerging role of non-coding RNA in health and disease. Metab Brain Dis 2021; 36(6):1119-34. doi: 10.1007/s11011-021-00739-y [Crossref] [ Google Scholar]
- Arasaratnam D, Bell KM, Sim CB, Koutsis K, Anderson DJ, Qian EL. The role of cardiac transcription factor NKX2-5 in regulating the human cardiac miRNAome. Sci Rep 2019; 9(1):15928. doi: 10.1038/s41598-019-52280-9 [Crossref] [ Google Scholar]
- Fang X, Miao S, Yu Y, Ding F, Han X, Wu H. MIR148A family regulates cardiomyocyte differentiation of human embryonic stem cells by inhibiting the DLL1-mediated NOTCH signaling pathway. J Mol Cell Cardiol 2019; 134:1-12. doi: 10.1016/j.yjmcc.2019.06.014 [Crossref] [ Google Scholar]
- Zhao H, Choi K. Single cell transcriptome dynamics from pluripotency to FLK1( + ) mesoderm. Development 2019; 146(23):dev182097. doi: 10.1242/dev.182097 [Crossref] [ Google Scholar]
- Meng Z, Wang J, Peng J, Zhou Y, Zhou S, Song W. Dynamic transcriptome profiling toward understanding the development of the human embryonic heart during different Carnegie stages. FEBS Lett 2020; 594(24):4307-19. doi: 10.1002/1873-3468.13930 [Crossref] [ Google Scholar]
- Mononen MM, Leung CY, Xu J, Chien KR. Trajectory mapping of human embryonic stem cell cardiogenesis reveals lineage branch points and an ISL1 progenitor-derived cardiac fibroblast lineage. Stem Cells 2020; 38(10):1267-78. doi: 10.1002/stem.3236 [Crossref] [ Google Scholar]
- Yamak A, Hu D, Mittal N, Buikema JW, Ditta S, Lutz PG. Loss of Asb2 impairs cardiomyocyte differentiation and leads to congenital double outlet right ventricle. iScience 2020; 23(3):100959. doi: 10.1016/j.isci.2020.100959 [Crossref] [ Google Scholar]
- Pezhouman A, Nguyen NB, Sercel AJ, Nguyen TL, Daraei A, Sabri S. Transcriptional, electrophysiological, and metabolic characterizations of hESC-derived first and second heart fields demonstrate a potential role of TBX5 in cardiomyocyte maturation. Front Cell Dev Biol 2021; 9:787684. doi: 10.3389/fcell.2021.787684 [Crossref] [ Google Scholar]
- Yang D, Gomez-Garcia J, Funakoshi S, Tran T, Fernandes I, Bader GD, et al. Modeling human multi-lineage heart field development with pluripotent stem cells. Cell Stem Cell 2022;29(9):1382-401.e8. 10.1016/j.stem.2022.08.007.
- Zawada D, Kornherr J, Meier AB, Santamaria G, Dorn T, Nowak-Imialek M. Retinoic acid signaling modulation guides in vitro specification of human heart field-specific progenitor pools. Nat Commun 2023; 14(1):1722. doi: 10.1038/s41467-023-36764-x [Crossref] [ Google Scholar]
- Guo H, Hang C, Lin B, Lin Z, Xiong H, Zhang M. HAND factors regulate cardiac lineage commitment and differentiation from human pluripotent stem cells. Stem Cell Res Ther 2024; 15(1):31. doi: 10.1186/s13287-024-03649-9 [Crossref] [ Google Scholar]
- Lu SH, Lee KZ, Hsu PW, Su LY, Yeh YC, Pan CY. Alternative splicing mediated by RNA-binding protein RBM24 facilitates cardiac myofibrillogenesis in a differentiation stage-specific manner. Circ Res 2022; 130(1):112-29. doi: 10.1161/circresaha.121.320080 [Crossref] [ Google Scholar]
- Henry S, Szabó V, Sutus E, Pirity MK. RYBP is important for cardiac progenitor cell development and sarcomere formation. PLoS One 2020; 15(7):e0235922. doi: 10.1371/journal.pone.0235922 [Crossref] [ Google Scholar]
- Cho SW, Kim HK, Sung JH, Kim Y, Kim JH, Han J. Mitochondrial energy metabolic transcriptome profiles during cardiac differentiation from mouse and human pluripotent stem cells. Korean J Physiol Pharmacol 2022; 26(5):357-65. doi: 10.4196/kjpp.2022.26.5.357 [Crossref] [ Google Scholar]
- Hansel-Frose AF, Allmer J, Friedrichs M, Dos Santos HG, Dallagiovanna B, Spangenberg L. Alternative polyadenylation and dynamic 3’ UTR length is associated with polysome recruitment throughout the cardiomyogenic differentiation of hESCs. Front Mol Biosci 2024; 11:1336336. doi: 10.3389/fmolb.2024.1336336 [Crossref] [ Google Scholar]
- Miklas JW, Clark E, Levy S, Detraux D, Leonard A, Beussman K. TFPa/HADHA is required for fatty acid beta-oxidation and cardiolipin re-modeling in human cardiomyocytes. Nat Commun 2019; 10(1):4671. doi: 10.1038/s41467-019-12482-1 [Crossref] [ Google Scholar]
- Chen X, Liu Y, Xu C, Ba L, Liu Z, Li X. QKI is a critical pre-mRNA alternative splicing regulator of cardiac myofibrillogenesis and contractile function. Nat Commun 2021; 12(1):89. doi: 10.1038/s41467-020-20327-5 [Crossref] [ Google Scholar]
- Hang C, Song Y, Li Y, Zhang S, Chang Y, Bai R. Knockout of MYOM1 in human cardiomyocytes leads to myocardial atrophy via impairing calcium homeostasis. J Cell Mol Med 2021; 25(3):1661-76. doi: 10.1111/jcmm.16268 [Crossref] [ Google Scholar]
- Hoffmann S, Schmitteckert S, Raedecke K, Rheinert D, Diebold S, Roeth R. Network-driven discovery yields new insight into Shox2-dependent cardiac rhythm control. Biochim Biophys Acta Gene Regul Mech 2021; 1864(4-5):194702. doi: 10.1016/j.bbagrm.2021.194702 [Crossref] [ Google Scholar]
- Rohner E, Witman N, Sohlmer J, De Genst E, Louch WE, Sahara M. An mRNA assay system demonstrates proteasomal-specific degradation contributes to cardiomyopathic phospholamban null mutation. Mol Med 2021; 27(1):102. doi: 10.1186/s10020-021-00362-8 [Crossref] [ Google Scholar]