Biomed adv. 2(3):164-170.
doi: 10.34172/bma.27
Original Article
Downregulation of miR-372 in non-proliferating human pluripotent stem cell derived cardiomyocytes
Mahshad Shiri Formal analysis, Methodology, 1 
Fatemeh Etezadi Formal analysis, Methodology, 1
Seyed Parham Hosseini Writing – original draft, 1
Sedigheh Gharbi Formal analysis, Methodology, 2
Sara Pahlavan Conceptualization, Funding acquisition, Supervision, Validation, Writing – review & editing, 1, * 
Author information:
1Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
2Department of Biology, Faculty of Science, Shahid Bahonar University of Kerman, Kerman, Iran
Abstract
Summary
Introduction:
The cell cycle arrest of the mammalian adult cardiomyocytes is the main reason for limited cardiac regeneration. A complex network of intracellular molecules facilitates cell cycle progression, from which regulatory proteins are well studied. However, the non-protein compartments such as regulatory microRNAs (miRNAs) are underrepresented. Here, we explored the miRNAs with differential expression in proliferating and non-proliferating cardiomyocytes.
Methods:
Candidate miRNAs with significant differential expression between 14-day and 45-day human embryonic stem cell-derived cardiomyocytes (hESC-CM) were identified using reanalysis of data set GSE35672. Human embryonic stem cells (hPSCs) were expanded and differentiated into cardiomyocytes by a cocktail of small molecules targeting Wnt/β-catenin and TGF-β signaling, and samples were collected for expression analysis of in silico-identified candidate miRNAs at days 10, 20, and 30 of differentiation.
Findings:
miR-302d, miR-371-5p, and miR-372 were selected as candidate differentially expressed miRNAs (DEmiRNAs). While miR-302d and miR-371-5p expression did not repeat the in-silico results in cTNT+hESC-CM, miR-372 showed a significant downregulation from day 10 to day 30.
Conclusion:
This finding suggests a possible regulatory effect of miRNA-372 in cell cycle arrest of mature cardiomyocytes.
Keywords: Human embryonic stem cell, Cardiomyocyte, Cell cycle, microRNA
Copyright and License Information
© 2025 The Author(s).
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Funding Statement
This work was supported by a grant from Iran National Science Foundation (grant number: 4020198).
Introduction
Cardiovascular diseases comprise a major number of mortality worldwide. According to the Global Burden of Disease (GBD) Study 2019, 18.6 million people died from cardiovascular diseases in this year.1 This statistic warns us about finding more efficient and appropriate treatments and manage the spread of cardiovascular disorders.Despite efforts to advance treatment methods in recent decades, there is still a long way to go in order to achieve a significant improvement.
Ischemic heart diseases are the most common known cardiovascular disorder, caused by plaque formation in the coronary arteries that supply nutrients and oxygen to the myocardium. The cessation or reduction of blood flow to the myocardium, due to coronary artery occlusion, causes a heart attack. This process leads to the destruction and death of heart muscle cells, or cardiomyocytes.2 Cardiomyocyte proliferation is a key factor in the repair of cardiac tissue following injury. Research has shown that mammalian cardiomyocytes lose their ability to divide after birth due to terminal differentiation and exit from the cell cycle. Unlike mammals, some fish, reptiles, and amphibians maintain the cardiomyocytes’ proliferation capacity throughout life.This difference originates from various growth pathways that are employed in different organisms. As a result, targeting genes involved in cell division could be a strategy to restore the ability of repair in the heart after injury.3
The cell cycle is controlled by two groups of proteins; cyclins and cyclin-dependent kinases (CDKs). The activity of cyclins and CDKs is regulated by two families of CDK inhibitors, INK4s and CIP/KIPs.These inhibitors control cell cycle by binding to CDKs and preventing the formation of the cyclin-CDK complex.4 In addition to being regulated by a complex network of proteins, recent studies have shown that non-coding RNAs has a considerable role in cell cycle regulation by influencing various processes, including protein expression. Porrello and colleagues discovered a key regulatory role for the microRNA-15 (miR-15) family in the mitotic arrest of cardiomyocytes after birth 5. Initially, they identified a list of miRNAs which showed differential expression at postnatal days 1 and 10 in mice and narrowed down their list to the miR-15 family, particularly miR-195 with highest differential expression.Huang and colleagues investigated the role of miR-128 in cardiomyocytes’ proliferation and cardiac regeneration.6 Wang et al profiled miRNAs in the early phases of cardiomyocytes differentiation and found miR-25 with a significant differential expression in proliferating and non-proliferating human pluripotent stem cells-derived cardiomyocytes (hPSC-CMs).7 Borden et al introduced miR-294 as a regulator of cardiomyocytes cell cycle re-entry.8 Bian and colleagues could enhance myocardial repair by overexpressing miR-199a in cardiomyocytes derived from hPSCs.9 In the current study, we reanalyzed a public dataset for miRNA expression in early and late phases of cardiomyocytes differentiation and explored the in-silico identified miRNAs in proliferating and non-proliferating hPSC-CMs.
Methods
Dataset selection and RNA-seq data reanalysis
In order to find public datasets for miRNA expression in immature (proliferating) and mature (non-proliferative) cardiomyocytes, we used the following keywords in NCBI; cardiomyocytes, cell cycle arrest and miRNAs. The result was a large number of datasets, from which we selected one that resembled Royan in vitro platform of hPSC-CM differentiation (GSE35672).10 This selected dataset studied miRNA transcripts in differentiating human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes from day 0 to 120 of differentiation. The data related to days 10 (immature cardiomyocytes) and 45 (mature cardiomyocytes) were reanalyzed using the GEO2R Web tool with settings adjusted P value < 0.05 and Log2 Fold change > |2|. MirTarBase and EnrichR were used to explore the target genes and signaling pathways, respectively. The overview of workflow is depicted in Figure 1.
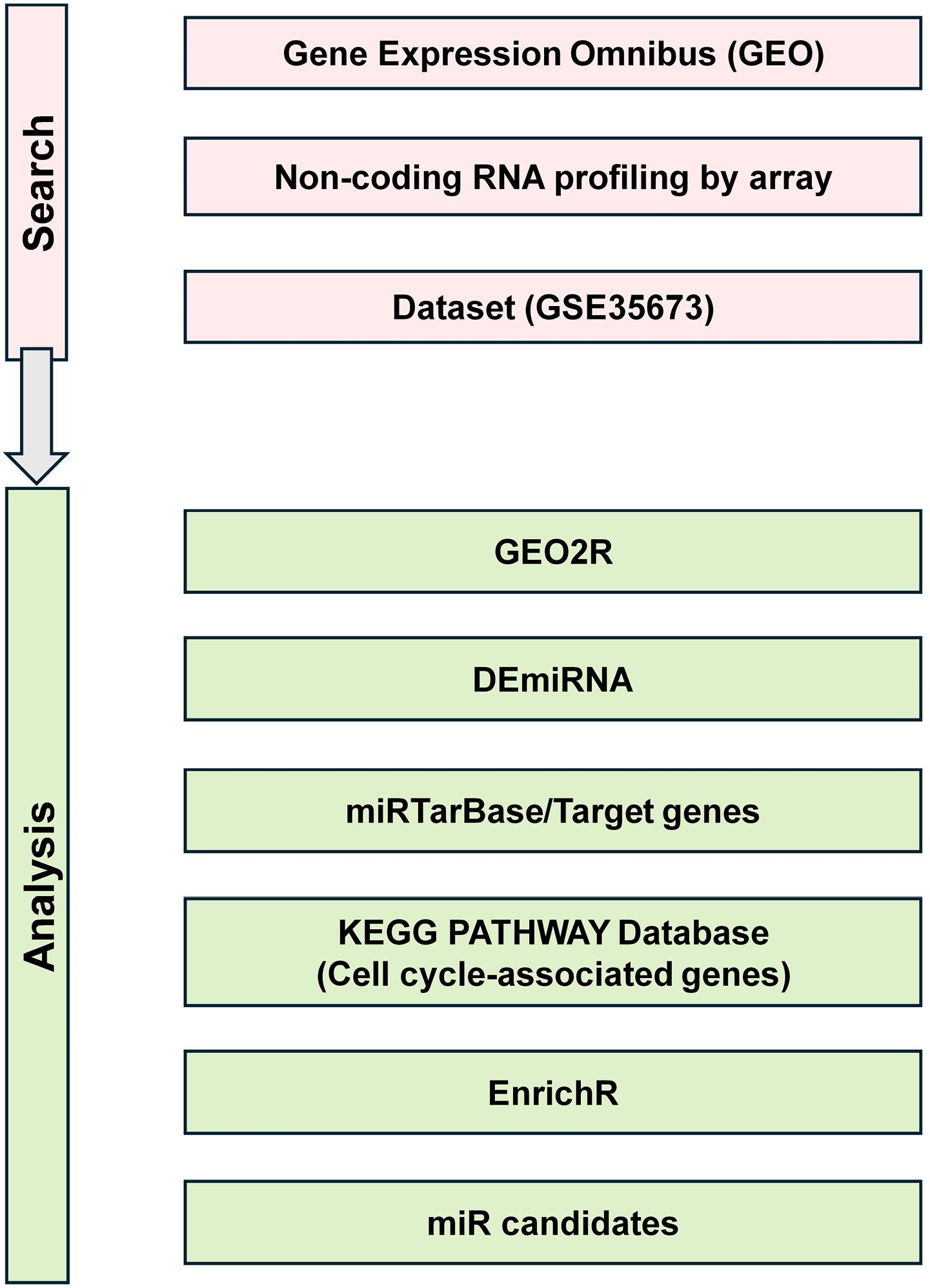
Figure 1.
Workflow of transcriptomic data reanalysis for identification of cardiomyocytes cell cycle–associated miRNAs
.
Workflow of transcriptomic data reanalysis for identification of cardiomyocytes cell cycle–associated miRNAs
hESCs expansion and differentiation into cardiomyocytes
Human embryonic stem cells (Royan H6 [RH6] line) were obtained from the Royan Institute’s Stem Cell Bank, expanded and differentiated as follow: induction towards mesendoderm using small molecule (SM) CHIR (12 µM, Stemgent, 04-0004-10), towards cardiac mesoderm and cardiac progenitor cells using 5 µM of 3 SMs IWP2 (Tocris Bioscience, 3533), SB431542 (Cayman, 13031) and purmorphamine (Stemgent, 04–0009). Spontaneous beating was observed by day 7 of differentiation (D7). The human embryonic stem cells-derived cardiomyocytes (hESC-CM) was maintained in culture until day 30 of differentiation (D30) and cardiomyocytes were collected at days 10, 20 and 30 (D10, D20, D30) for assessments.
Immunostaining
To assess cardiomyocyte specification at protein level, immunostaining was performed for cardiac troponin T (cTNT). Cells were fixed using 4% paraformaldehyde for 15 minutes, permeabilized with 0.1% Triton in PBS for 20 minutes, blocked with 1% BSA for 45 minutes, incubated with primary antibody (Abcam, AB6423) for 2 hours at room temperature (RT) and secondary antibody (Donkey anti-goat, Abcam, AB150132) for 1 hour at 37 °C. For nuclear staining, DAPI (Sigma-Aldrich, D8417) was used for 30 seconds. Images were captured using an Olympus IX71 fluorescence microscope.
RNA isolation and quantitative RT-PCR (qRT -PCR)
hESC and D10, D20 and D30 hESC-CM were collected for RNA extraction. RNA isolation was done using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. To remove any potential DNA contamination, samples were incubated with RNase-free DNase (Takara, Japan). The extracted RNAs were elongated using the Poly(A) Polymerase Tailing Kit from Ratin Gene. The resultant elongated RNA was reverse-transcribed into cDNA and diluted to 50 ng/μL for qRT-PCR in the Rotor Gene 6000 (Corbett, Australia). The analysis was carried out using REST analysis software (QIAGEN, Germany). U48 was used as the housekeeping gene. The primer sequences of miRNAs are available in Supplementary file, Table S1.
Statistical analysis
Data are presented as mean ± SD/SEM from at least three biological replicates. Statistical significance was assessed using one-way Analysis of Variance (ANOVA) in GraphPad Prism software (GraphPad Software, USA). *P < 0.05, ** P < 0.01, and *** P < 0.001 were considered as statistically significant.
Results
miRNAs with differential expression at early and late differentiation days
First, we searched the GEO database for the keywords miRNA, cardiomyocyte and cell cycle arrest. The initial survey resulted in several datasets from which GSE35673 was selected due to similar in vitro cardiomyocyte differentiation platform. To compare immature (proliferating) and mature (non-proliferating) hESC-CM, we picked days 14 and 45 of differentiation because in vitro cardiomyocyte differentiation protocols result into more mature cardiac cells by extending culture days. We could use day 120 as mature cardiomyocytes, but day 45 was used to be equivalent to day 30 of our protocol which showed non proliferating cells in our previous study.11 The samples related to days 14 and 45 was assessed with respect to normalization status (Figure 2A) which confirmed suitable data distribution. Furthermore, uniform manifold approximation and projection (UMAP) that estimates a topology of the high-dimensional data by a non-linear dimensionality reduction method, was used which results reflected the proximity of data in the replicates of control (day 10) and test (day 45) (Figure 2B). Differentially expressed miRNAs were identified, which are listed in Supplementary file 2 and visualized in Volcano plot (Figure 2C) and Venn diagram (Figure 2D). Based on adjusted p value < 0.05 and Log2 Fold change > |2|, significantly upregulated and downregulated miRNAs were selected and listed in Table 1.
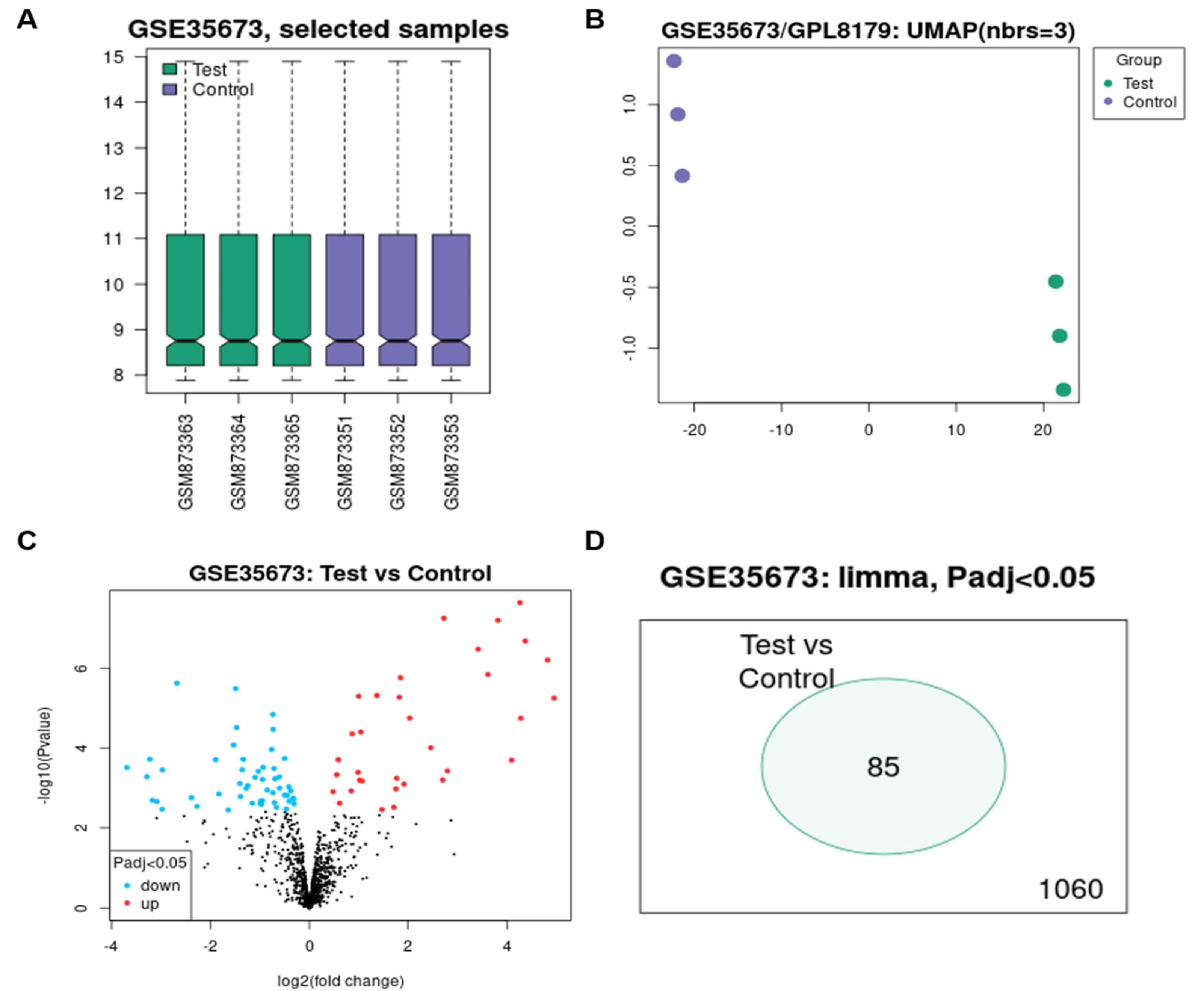
Figure 2.
Visualization of bioinformatics analysis. (A) BoxPlot related to the analyzed data, which includes three repeats of control (D10) and three repeats of test (D45), and shows a uniform distribution of data all over control and test repeats. (B) UMAP plot showing complete separation of control and test data. (C) Volcano plot showing upregulated and downregulated miRNAs in black and those with significant expression changes in red (Up) and blue (down). (D) Venn diagram showing 85 common miRNAs with expression changes between control and test data
.
Visualization of bioinformatics analysis. (A) BoxPlot related to the analyzed data, which includes three repeats of control (D10) and three repeats of test (D45), and shows a uniform distribution of data all over control and test repeats. (B) UMAP plot showing complete separation of control and test data. (C) Volcano plot showing upregulated and downregulated miRNAs in black and those with significant expression changes in red (Up) and blue (down). (D) Venn diagram showing 85 common miRNAs with expression changes between control and test data
Table 1.
Significantly upregulated and downregulated miRNAs
|
ID
|
Adjusted
P
value
|
P
value
|
logFC
|
miRNA_ID
|
|
Down-regulated miRNAs
|
| ILMN_3167463 |
0.0108167 |
3.02E-04 |
-3.69027917 |
hsa-miR-373 |
| ILMN_3167164 |
0.0145043 |
5.11E-04 |
-3.28807111 |
hsa-miR-371-3p |
| ILMN_3167547 |
0.0075609 |
1.87E-04 |
-3.22926361 |
hsa-miR-122 |
| ILMN_3167229 |
0.0332343 |
1.99E-03 |
-3.17575333 |
hsa-miR-375 |
| ILMN_3168586 |
0.0338695 |
2.13E-03 |
-3.08460139 |
hsa-miR-371-5p |
| ILMN_3166944 |
0.0458151 |
3.32E-03 |
-2.97823111 |
hsa-miR-302c |
| ILMN_3168322 |
0.0113157 |
3.46E-04 |
-2.97399056 |
hsa-miR-302b* |
| ILMN_3168595 |
0.0003018 |
2.37E-06 |
-2.67872278 |
hsa-miR-302d* |
| ILMN_3167474 |
0.0300407 |
1.71E-03 |
-2.384 |
hsa-miR-302a* |
| ILMN_3167386 |
0.040757 |
2.81E-03 |
-2.2723 |
hsa-miR-302d |
|
Up-regulated miRNAs
|
| ILMN_3167551 |
0.0004547 |
5.56E-06 |
4.9517475 |
hsa-let-7d |
| ILMN_3168365 |
0.0001183 |
6.20E-07 |
4.82217556 |
hsa-let-7g |
| ILMN_3168316 |
0.0000595 |
2.08E-07 |
4.36871 |
hsa-let-7i |
| ILMN_3167189 |
0.0011915 |
1.77E-05 |
4.27729861 |
hsa-let-7f |
| ILMN_3167970 |
0.0000243 |
2.30E-08 |
4.25894583 |
hsa-let-7b |
| ILMN_3167422 |
0.0075609 |
1.98E-04 |
4.09181 |
hsa-miR-98 |
| ILMN_3168513 |
0.0000243 |
6.36E-08 |
3.81552944 |
hsa-let-7c |
| ILMN_3167971 |
0.0002337 |
1.43E-06 |
3.61442222 |
hsa-let-7a |
| ILMN_3168710 |
0.0000765 |
3.34E-07 |
3.41608167 |
hsa-let-7d* |
| ILMN_3167105 |
0.0116751 |
3.67E-04 |
2.79325278 |
hsa-miR-208b |
| ILMN_3168724 |
0.0000243 |
5.65E-08 |
2.72172722 |
hsa-let-7i* |
| ILMN_3167652 |
0.0151889 |
6.17E-04 |
2.69868917 |
hsa-miR-452*:9.1 |
| ILMN_3168180 |
0.0048193 |
9.68E-05 |
2.45632778 |
hsa-miR-378* |
| ILMN_3168711 |
0.0011915 |
1.76E-05 |
2.02899778 |
hsa-let-7e* |
Blue color shows downregulated miRNAs and orange color depicts upregulated ones.
The downregulated miRNAs were used in the rest of the in silico analysis, because we aimed to introduce regulatory miRNAs for cell cycle re-entry by gene overexpression systems. The mRNA targets of downregulated miRNAs were identified using MirTarBase and the cell cycle-related genes were determined by comparing target mRNAs to the list of cell cycle genes obtained from KEGG (Supplementary file 3). Furthermore, the associated signaling pathways of cell cycle-related target mRNAs (such as CUL1, TAOK1, FASLG and APC2) were determined using Enrichr as (i) Pathway in cancer, (ii) Wnt signaling pathway, (iii) Hedgehog signaling pathway (iv) MAPK signaling pathway, (v) Thyroid hormone signaling pathway (vi) Dilated cardiomyopathy (Table 2).
Table 2.
Signaling pathways for cell cycle-related target mRNAs
|
Signaling pathway
|
Target mRNA
|
| Pathway in cancer |
ITGA6 |
| PRKACB |
| APC2 |
| COL1 |
| FASLG |
| Wnt signaling pathway |
APC2 |
| PRKACB |
| COL1 |
| MAPK signaling |
FASLG |
| PRKACB |
| TAOK1 |
| hedgehog signaling pathway |
COL1 |
| PRKACB |
| Thyroid hormone signaling pathway |
PRKACB |
| THRB |
| Dilated cardiomyopathy |
ITGA6 |
| PRKACB |
Considering the frequency of cell cycle-related genes among the target mRNAs of candidate miRNAs, we categorized them in the following order: (1) miR-302: 21 cell cycle-related target mRNAs, (2) miR-373: 10 (3) miR-371: 10, (4) miR-375: 5, (5) miR-122: 0.
Because-miR-122 did not have any target mRNAs related to the cell cycle, it was removed from our list and 4 groups of miRNAs including group 1 (miR-302a, miR-302b, miR-302c, miR-302d), group 2 (miR-373), group 3 (miR-371-3p, miR-371-5p) and group 4 (miR-375) were selected for further studies. During primer design for candidate miRNAs of group 1 to 4, we found abundant sequences between candidates which limited the possibility of specific primer designs for each candidate. Thus, we substituted miRNA-373 and miRNA-375 with miRNA-372. The miRNA-372 did not shortlist in our initial analysis because of adjusted p-value. However, Log2 FC was greater than -2. Ultimately, primers were designed for three candidates including miR-302d, miR-371-5p and miR-372 (Table S1).
In vitro cardiomyocyte differentiation resembling heart development
The adherent culture was employed to expand hESCs. Figure 3A represents a brief overview of differentiation protocol including static suspension culture for hESC spheroid formation prior to differentiation, one day CHIR treatment for mesendoderm induction, and two days of 3 SMs (IWP2, SB and purmorphamine) for cardiac progenitor cell derivation. Spontaneously beating cardiomyocytes were observed at day 7 of differentiation (D7) (Figure 3A). While the NKX2.5+cells were the major population at day 4 cardiac progenitor cells, cardiomyocytes expressed cTNT widely (Figure 3B).
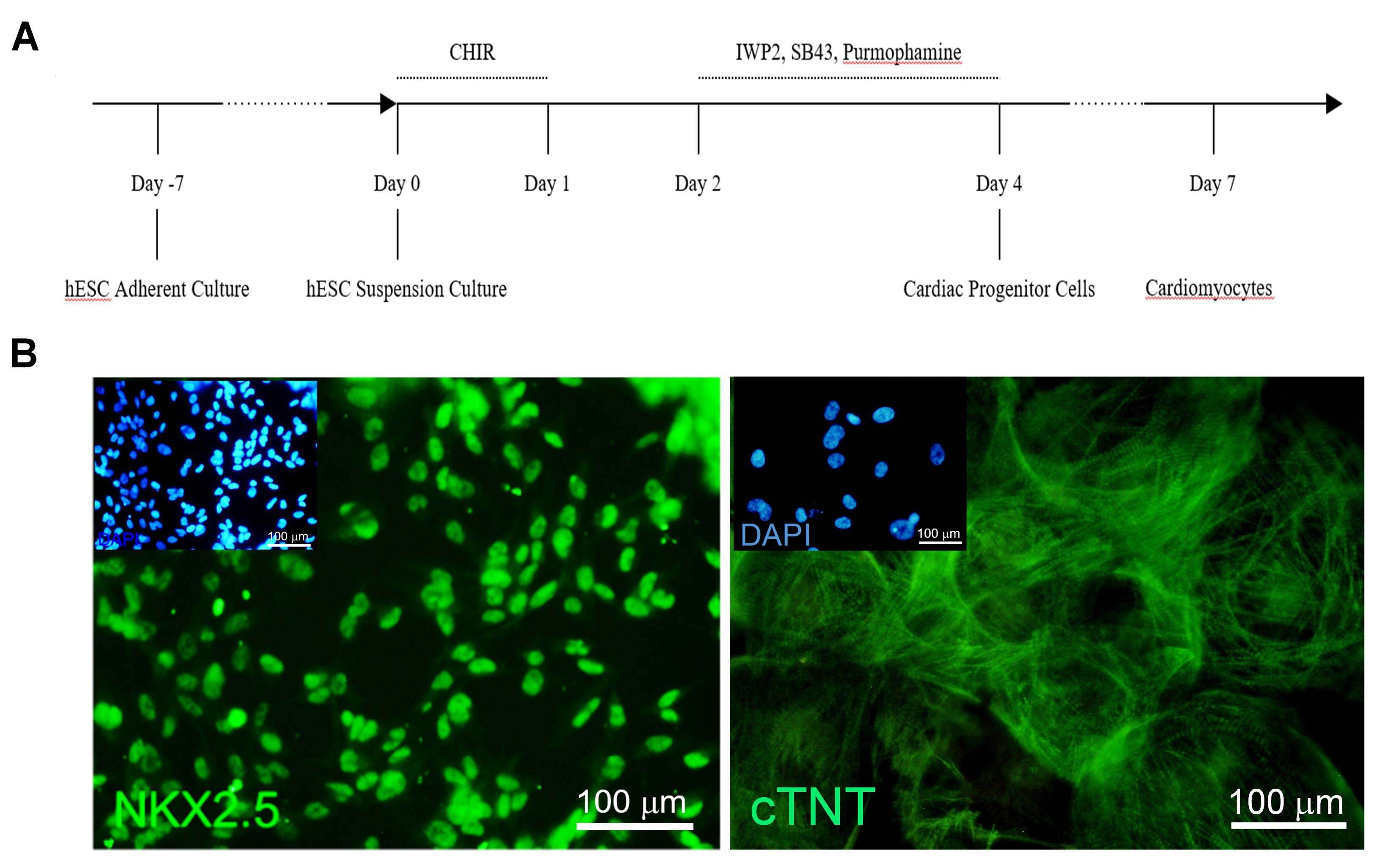
Figure 3.
Cardiomyocyte differentiation and characterization (A) Schematic diagram of differentiation protocol. (B) Immunofluorescence staining showing NANOG expression at D0 hESCs and cardiac troponin T (cTNT) expression at D30 of cardiomyocyte differentiation. ‘D’ day of differentiation
.
Cardiomyocyte differentiation and characterization (A) Schematic diagram of differentiation protocol. (B) Immunofluorescence staining showing NANOG expression at D0 hESCs and cardiac troponin T (cTNT) expression at D30 of cardiomyocyte differentiation. ‘D’ day of differentiation
Expression analysis of candidate miRNAs in differentiated cardiomyocytes
We collected cardiomyocytes at D0, D10, D20 and D30 of differentiation and assessed the expression of candidate miRNAs. As shown in Figure 4A, the expression of miRNA-302d decreased at D0, but showed an increasing trend by extending culture days to D20 and D30. miRNA-371-5p expression did not change at D10 compared to D0, but increased at D20 and D30 (Figure 4B). While the expression of miRNA-372 markedly increased at D10 of differentiation, a significant expression decrease was observed at days 20 and 30. The significant downregulation of miRNA-372 from D10 to D30 resembled the in silico prediction (Figure 4C).
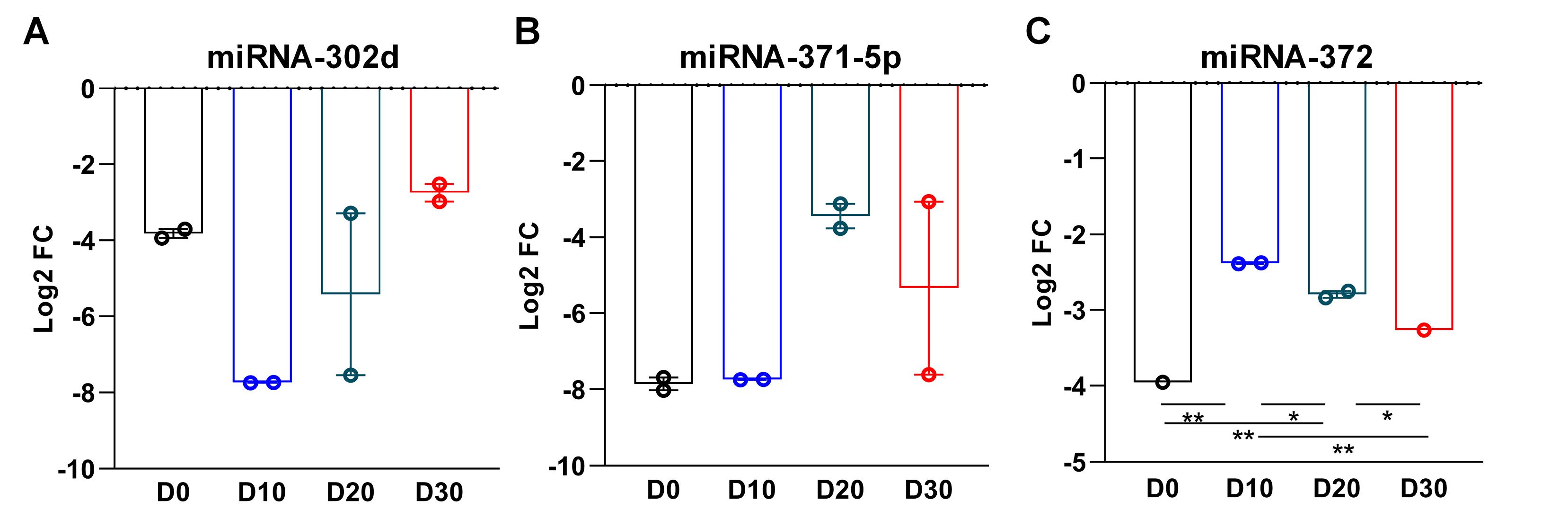
Figure 4.
Expression analysis of candidate miRNAs in hESC-derived cardiomyocytes. qRT-PCR analysis comparing the expression of three miRNA candidates (miRNA-302d, miRNA-371-5p and miRNA-372) at days 0 D0), 10 (D10), 20 (D20) and 30 (D30) of differentiation. miRNA-372 was significantly downregulated from D10 to D30, being coherent with the in silico predictions. Data are presented as mean ± SEM (n = 3); *P < 0.05, ** P < 0.01 (One-way ANOVA)
.
Expression analysis of candidate miRNAs in hESC-derived cardiomyocytes. qRT-PCR analysis comparing the expression of three miRNA candidates (miRNA-302d, miRNA-371-5p and miRNA-372) at days 0 D0), 10 (D10), 20 (D20) and 30 (D30) of differentiation. miRNA-372 was significantly downregulated from D10 to D30, being coherent with the in silico predictions. Data are presented as mean ± SEM (n = 3); *P < 0.05, ** P < 0.01 (One-way ANOVA)
Discussion
The mammalian cardiomyocytes undergo cell cycle arrest after birth which limit the cardiac regeneration and repair after injury.3 To investigate the unknown molecular mechanisms of this process, we checked differentially expressed miRNAs in proliferating and non-proliferating hESC-derived cardiomyocytes. The common result of in silico and in vitro assays identified miRNA-372 as a downregulated miRNA, which might have a role in the regulation of cell cycle arrest.
The cell cycle is a vital biological process, which supports growth and regeneration.3 Some cell types lose their proliferation ability after birth including cardiomyocytes and neurons.3 Several studies investigated the regulatory proteins involved in this process.12 However, the regulatory RNAs have been less studied. The microRNAs may regulate the cell cycle post-transcriptionally.13
The miR-371-5p is in a cluster located on human chromosome 19 and plays multiple regulatory roles, including a rise in cancer cell proliferation and tumor growth as well as regulating immune responses, post-transcriptional processes, and cardiomyocyte proliferation.14 It has been shown that miRNA-371-5p promotes tumor growth by regulating the ZNF749 gene in pancreatic cancer cells.15 It also affects the activity of immune cells such as macrophages and T cells and regulates pathological processes such as inflammation and fibrosis.16 In myocardium, it increases cardiomyocyte proliferation by targeting the LATS1 and LATS2 in the Hippo pathway.17
miR-302d is located on human chromosome 4 and has regulatory roles in various human cell types, including maintaining the stemness state of embryonic stem cells, regulating the deletion of maternal transcripts, and influencing the expression of important genes in the stem cells such as OCT4 and NANOG.18 One study demonstrated that miR-302d-3p can regulate the survival, migration, and apoptosis of breast cancer cells through the regulation of the TMBIM6-mediated ERK signaling pathway.19
miR-372 is located on human chromosome 19 and has multiple regulatory roles in various human cell types, including regulating cancer cell proliferation by reducing LATS2 gene expression, regulating the cell cycle, and preventing uncontrolled cell proliferation.20 It regulates apoptosis and immune responses by influencing the activity of immune cells such as macrophages and T cells, and regulates post-transcriptional processes to maintain homeostasis and function of various cells.21 It was shown that dysregulation of miR-372-5p expression plays an important role in the development of gastric cancer.22 Furthermore, upregulation of miR-372-5p increased CDX2 and decreased CDX1 expression, while inhibition of miR-372-5p reversed this expressional change.23
Among the three miRNA candidates, only miRNA-372 resembled the in silico expression pattern. This controversy might originate from a slightly different differentiation protocol used for generation of GSE35672 dataset,lack of a selection step and heterogeneity of cell population.
Conclusion
In conclusion, this study introduced miRNA-372 as a possible regulator of cardiomyocyte cell cycle arrest. Given that post-natal cardiomyocytes stop the cell cycle, the heart regeneration faces a significant challenge. Therefore, it is imperative to gain knowledge on the regulators of this process on multiple levels of proteins and RNAs, in order to re-stablish the cell cycle after cardiomyocyte injury.
Competing Interests
The authors declare no conflicts of interest.
Ethical Approval
The ethical approval was not required for the current study as investigated and issued by the Royan Institute Ethics Committee under the license number IR.ACECR.ROYAN.REC.1402.063 (October 7th, 2023). Because the hESC line which was used in this study (RH6), was derived in the project entitled “Generation of new human embryonic stem cell lines with diploid and triploid karyotypes”. The project was performed following the approval of Royan Institute Ethics Committee and after obtaining informed consent from the couple undergoing in vitro fertilization treatment (2006, full information can be found in https://hpscreg.eu/cell-line/RIe006-A).24
Intelligence Use Disclosure
This article has not utilized artificial intelligence (AI) tools for research and manuscript development, as per the GAMER reporting guideline.
Supplementary Files
Supplementary file 1 contains Table S1 (List of primers used for qRT-PCR).
(pdf)
Supplementary file 2. RNAseq analysis output of differentially expressed miRNAs.
(xlsx)
Supplementary file 3. mRNA targets of significantly downregulated miRNAs.
(pdf)
Acknowledgements
The authors would like to thank all members of Royan Cardiovascular Group for the help and support.
References
- Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM. Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J Am Coll Cardiol 2020; 76(25):2982-3021. doi: 10.1016/j.jacc.2020.11.010 [Crossref] [ Google Scholar]
- Chiong M, Wang ZV, Pedrozo Z, Cao DJ, Troncoso R, Ibacache M. Cardiomyocyte death: mechanisms and translational implications. Cell Death Dis 2011; 2(12):e244. doi: 10.1038/cddis.2011.130 [Crossref] [ Google Scholar]
- Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev 2007; 87(2):521-44. doi: 10.1152/physrev.00032.2006 [Crossref] [ Google Scholar]
- Tane S, Okayama H, Ikenishi A, Amemiya Y, Nakayama KI, Takeuchi T. Two inhibitory systems and CKIs regulate cell cycle exit of mammalian cardiomyocytes after birth. Biochem Biophys Res Commun 2015; 466(2):147-54. doi: 10.1016/j.bbrc.2015.08.102 [Crossref] [ Google Scholar]
- Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res 2011; 109(6):670-9. doi: 10.1161/circresaha.111.248880 [Crossref] [ Google Scholar]
- Huang W, Feng Y, Liang J, Yu H, Wang C, Wang B. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat Commun 2018; 9(1):700. doi: 10.1038/s41467-018-03019-z [Crossref] [ Google Scholar]
- Wang B, Xu M, Li M, Wu F, Hu S, Chen X. miR-25 promotes cardiomyocyte proliferation by targeting FBXW7. Mol Ther Nucleic Acids 2020; 19:1299-308. doi: 10.1016/j.omtn.2020.01.013 [Crossref] [ Google Scholar]
- Borden A, Kurian J, Nickoloff E, Yang Y, Troupes CD, Ibetti J. Transient introduction of miR-294 in the heart promotes cardiomyocyte cell cycle reentry after injury. Circ Res 2019; 125(1):14-25. doi: 10.1161/circresaha.118.314223 [Crossref] [ Google Scholar]
- Bian W, Chen W, Nguyen T, Zhou Y, Zhang J. miR-199a overexpression enhances the potency of human induced-pluripotent stem-cell-derived cardiomyocytes for myocardial repair. Front Pharmacol 2021; 12:673621. doi: 10.3389/fphar.2021.673621 [Crossref] [ Google Scholar]
- Babiarz JE, Ravon M, Sridhar S, Ravindran P, Swanson B, Bitter H. Determination of the human cardiomyocyte mRNA and miRNA differentiation network by fine-scale profiling. Stem Cells Dev 2012; 21(11):1956-65. doi: 10.1089/scd.2011.0357 [Crossref] [ Google Scholar]
- Ahmadvand S, Osia A, Meyfour A, Pahlavan S. Gender-specific characteristics of hypertrophic response in cardiomyocytes derived from human embryonic stem cells. J Cardiovasc Thorac Res 2021; 13(2):146-55. doi: 10.34172/jcvtr.2021.32 [Crossref] [ Google Scholar]
- Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res 2002; 90(10):1044-54. doi: 10.1161/01.res.0000020201.44772.67 [Crossref] [ Google Scholar]
- Zhu C, Yuan T, Krishnan J. Targeting cardiomyocyte cell cycle regulation in heart failure. Basic Res Cardiol 2024; 119(3):349-69. doi: 10.1007/s00395-024-01049-x [Crossref] [ Google Scholar]
- Li YJ, Dong M, Kong FM, Zhou JP, Liang D, Xue HZ. MicroRNA-371-5p targets SOX2 in gastric cancer. Oncotarget 2016; 7(22):31993-2005. doi: 10.18632/oncotarget.8289 [Crossref] [ Google Scholar]
- He D, Miao H, Xu Y, Xiong L, Wang Y, Xiang H. MiR-371-5p facilitates pancreatic cancer cell proliferation and decreases patient survival. PLoS One 2014; 9(11):e112930. doi: 10.1371/journal.pone.0112930 [Crossref] [ Google Scholar]
- Qiu YY, Zhang YW, Qian XF, Bian T. miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate Th1/Th2 balance in asthma through the combinatorial regulation of Runx3. Am J Transl Res 2017; 9(7):3184-99. [ Google Scholar]
- Song M, Wang H, Liu C, Jin S, Liu B, Sun W. Non-coding RNAs as regulators of the Hippo pathway in cardiac development and cardiovascular disease. Front Pharmacol 2024; 15:1348280. doi: 10.3389/fphar.2024.1348280 [Crossref] [ Google Scholar]
- Bourguignon LY, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem 2012; 287(39):32800-24. doi: 10.1074/jbc.M111.308528 [Crossref] [ Google Scholar]
-
Liao Y, Qiu Z, Bai L. miR-302d-3p regulates the viability, migration and apoptosis of breast cancer cells through regulating the TMBIM6-mediated ERK signaling pathway. Mol Med Rep 2021;24(6). doi: 10.3892/mmr.2021.12493.
- Meng Z, Li F, Wang B. miR-372-3p is a potential diagnostic factor for diabetic nephropathy and modulates high glucose-induced glomerular endothelial cell dysfunction via targeting fibroblast growth factor-16. Arch Med Sci 2023; 19(3):703-16. doi: 10.5114/aoms.2019.89659 [Crossref] [ Google Scholar]
- Tajik F, Alian F, Yousefi M, Azadfallah A, Hoseini A, Mohammadi F. MicroRNA-372 acts as a double-edged sword in human cancers. Heliyon 2023; 9(5):e15991. doi: 10.1016/j.heliyon.2023.e15991 [Crossref] [ Google Scholar]
- Zhou C, Li X, Zhang X, Liu X, Tan Z, Yang C. microRNA-372 maintains oncogene characteristics by targeting TNFAIP1 and affects NFκB signaling in human gastric carcinoma cells. Int J Oncol 2013; 42(2):635-42. doi: 10.3892/ijo.2012.1737 [Crossref] [ Google Scholar]
- Asghari Gharakhyli E, Tabar Molla Hassan A, Alipour M, Vahidi S, Samadani AA. The effect of miR-372-5p regulation on CDX1 and CDX2 in the gastric cancer cell line. Horm Mol Biol Clin Investig 2023; 44(3):271-6. doi: 10.1515/hmbci-2022-0045 [Crossref] [ Google Scholar]
- Baharvand H, Ashtiani SK, Taee A, Massumi M, Valojerdi MR, Yazdi PE. Generation of new human embryonic stem cell lines with diploid and triploid karyotypes. Dev Growth Differ 2006; 48(2):117-28. doi: 10.1111/j.1440-169X.2006.00851.x [Crossref] [ Google Scholar]